Researchers in East Africa add the Enterprise Breeding System to their work tools

Scientists overseeing breeding, principal technicians and data management and support staff from the International Maize and Wheat Improvement Center (CIMMYT) learned about the Enterprise Breeding System (EBS) at a training in Nairobi, Kenya, on May 4–6, 2022. This was the first in-person training on this advanced tool held in Eastern Africa.
Kate Dreher, Data Manager at CIMMYT, was the primary trainer. Dreher sought to ensure that scientists and their teams are well equipped to confidently use the EBS for their programs, including the creation and management of trials and nurseries. During the training, participants had the opportunity to test, review and give feedback on the system.
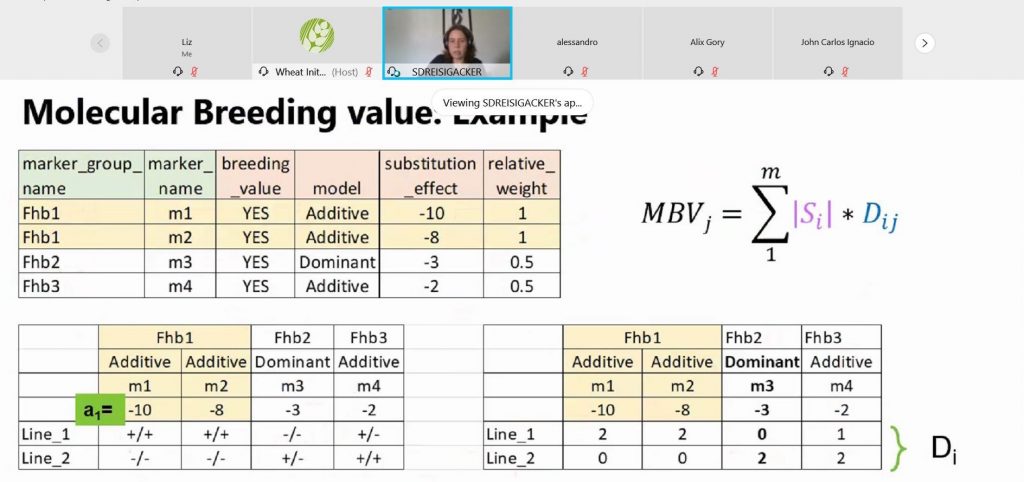
“The EBS is an online comprehensive system that brings together different types of data, including field observations and genotypic data, to harmonize processes across all teams and enable optimized decision-making in the short term and continuous learning for the long term,” Dreher said.
She explained that the EBS is more efficient than the former approach of using the Excel-based Maize Fieldbook software, even though it managed several useful processes.
The EBS is currently available to registered breeding and support team members and data managers from CIMMYT, IITA, IRRI and AfricaRice, across all geographies where related programs are implemented. Currently, the EBS is used by programs in maize, rice and wheat crops.
A more streamlined approach
“Although teams sent germplasm and phenotypic data for centralized storage in two databases (IMIS-GMS and MaizeFinder) managed by the data management team in Mexico in the past, this required curation after the data had already been generated,” Dreher said. “The EBS will enable teams to manage their germplasm and trial nursery data directly within one system.”
The EBS stores information on germplasm and linked seed inventory items. It is also designed to house and perform analyses using phenotypic and genotypic data. Users can also capture metadata about their trials and nurseries, such as basic agronomic management information and the GPS coordinates of sites where experiments are conducted.
Yoseph Beyene, Regional Maize Breeding Coordinator for Africa and Maize Breeder for Eastern Africa at CIMMYT, observed that the training gave him firsthand information on the current capabilities and use of the live version to search germplasm and seed, and the capabilities to create nurseries and trials.
“In the AGG project, we have one primary objective which focuses on implementing improved data management, experimental designs and breeding methods to accelerate genetic gain and improved breeding efficiency. Therefore, implementing EBS is one of the top priorities for AGG project,” said Yoseph, who leads the Accelerating Genetic Gains in Maize and Wheat for Improved Livelihoods Project (AGG).
Lourine Bii, an Assistant Research Associate who recently joined CIMMYT and the only female research technician on the Global Maize program based in Kenya, also found the training useful. “The EBS is a fantastic system that enables an individual to create experiments. The system links a team, for instance a product development team, to get live updates on the various stages of creating an experiment, reducing back and forth by email.”
The system’s software development is ongoing. The development team continues to add and enhance features based on feedback from users.