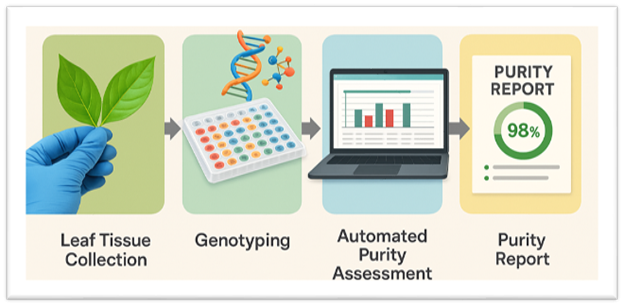
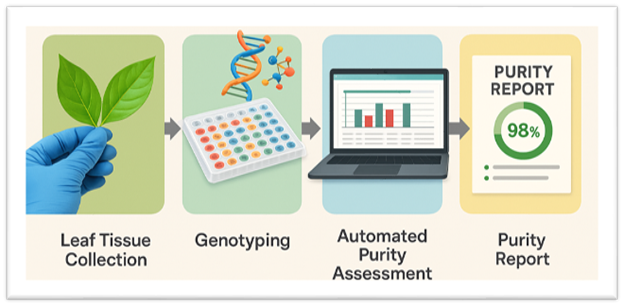
Fingerprinting bioinformatics pipeline cuts seed purity testing from days to clicks
A new bioinformatics-driven tool dramatically accelerates seed purity testing, offering fast, automated parental purity checks and hybridity verification—thereby increasing the effectiveness of breeders, seed companies, and regulators.
Waiting days—or even weeks—to analyze Single Nucleotide Polymorphism (SNP) assay data to verify the genetic purity and hybridity of seeds and crosses may soon be a thing of the past. A new bioinformatics-driven seed purity testing pipeline significantly reduces the time required to confirm seed identity, bringing it down to just a few clicks.
The new pipeline uses DNA fingerprinting with SNPs in a fast, accurate, and scalable automated genetic analysis.
Modernizing Genetic Purity and Hybridity Testing
Traditionally, seed producers and certification agencies have relied on grow-out tests and morphological analysis to assess genetic purity. These methods are labor-intensive and time-consuming, often requiring weeks or months of field or greenhouse work by trained technicians.
Although CIMMYT has long used molecular markers to verify parental purity and hybridity, challenges remained—particularly the lack of automated tools and integrated platforms to make the process seamless and accessible.
That is changing, thanks to software developed by Abhishek Rathore and team. The pipeline automatically compares each sample’s genetic profile to its expected reference using a custom algorithm. Based on user-defined thresholds, the tool confirms parental purity, identifies putative F1 hybrids, and flags failed crosses.
“We wanted a tool that breeders and seed companies can use without needing specialized bioinformatics skills,” said Abhishek Rathore, bioinformatics specialist at CIMMYT. “Once the DNA data is generated, the analysis is push-button. The software quickly interprets the SNP results and produces an easy-to-read report on seed purity. It’s about making advanced bioinformatics accessible and routine for parental purity and F1 verifications.”
Speedy, Automated, and User-Friendly
Early implementation of the pipeline has demonstrated large gains in speed and efficiency. What previously required extensive manual effort can now be completed in minutes.
The system is designed with user-friendliness in mind: lab technicians simply upload SNP assay results into an intuitive interface, and the pipeline returns clear metrics—such as “% purity”—while flagging any off-type individuals. With the computational workload fully automated, even seed companies and labs with minimal informatics infrastructure can benefit.
“Automation is key,” added Rathore. “By reducing manual steps and subjective interpretation, we save time and minimize human error. You can process dozens of seed samples overnight and receive a comprehensive genetic report by morning.”
To make this automation accessible to stakeholders across NARS, CIMMYT’s biometrician Roma Das developed a user-friendly web interface, while Peter Kimathi, a bioinformatics and software developer, developed a custom report and deployed the pipeline as a web service on CIMMYT’s servers (link below).
Widespread Adoption Across Africa
Since its rollout, the pipeline has been widely adopted by CIMMYT and partners through the Africa Dryland Crops Improvement Network (ADCIN). Mohan Chejerla, Genomics Expert at CIMMYT, has already applied the pipeline to over 23,000 samples, ensuring quality assurance and quality control (QA/QC) for breeding pipelines across Kenya, Uganda, Mali, Senegal, Burkina Faso, Ethiopia, Tanzania, Niger, Togo, Zambia, and Ghana.
This broad uptake underscores the demand for reliable, scalable seed purity testing—and the pipeline’s value for enhancing crop breeding and seed system integrity.
Additional Information:
🔗 Pipeline Source Code
🔗 CIMMYT Pipeline Implementation